Genetic diversity is critical for the health and genetic resilience of breeds. Litter Predict provides information for every planned mating comparing the litter to the selected parents and your breed as a whole to help you know if the match is going to preserve your breed’s rich genetic legacy. Read on to learn about how we measure diversity, why it matters, and some answers to frequently asked questions about this testing.
How we measure individual genetic diversity
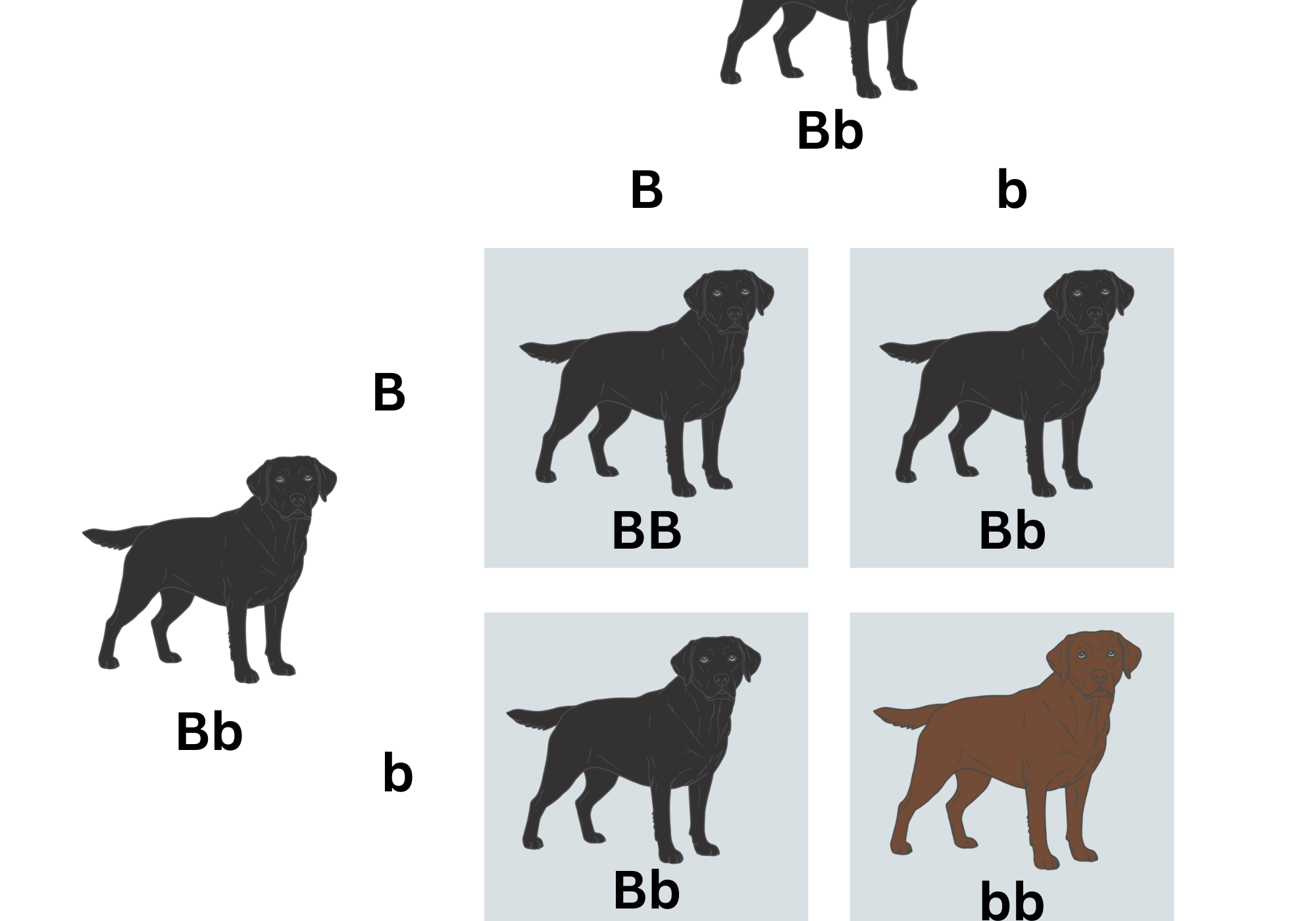
For each dog, we analyse thousands of specific markers selected for their ability to indicate genetic variation between dogs. These markers are not necessarily tied to visible variation between dogs, for example, coat colour. At any site, the DNA your dog inherits from each parent is either the same or different— scientists refer to these states as “homozygous” and “heterozygous,” respectively. The overall percentage of paired markers that are different is the percentage heterozygosity, or diversity, reported for each dog. The higher the heterozygosity, the more genetically diverse or outbred the dog is, and the farther to the right of the graph the dog will fall.
As each breed population has a unique history and gene pool, average diversity will vary by breed, and your dog’s percentage should be interpreted in light of its expected breed range. We provide dotted vertical lines indicating the typical range where the middle 80% of diversity scores for the breed lie (10-90th percentiles) based on breeder-tested dogs.

Why genetic diversity is important for breeders
Genetic diversity is often referred to as a ‘genetic toolbox’ that gives an organism options to respond to any challenge. What genetic variation each dog carries that makes them unique is largely unknown, but is nonetheless important to preserve in the breed “toolbox” for an uncertain future. As western studbooks did not fully close in most cases until the inter-war period, consideration of diversity is an increasingly real concern necessitated by concepts of breed and pedigree set in motion in the last one hundred years. A study in 2021 of over 200 purebred breeds showed that the average level of inbreeding (COI) in purebred dogs is around 0.25, or a heterozygosity in our testing of 33.1%, which is equivalent to the mating of full siblings. The effects of lost diversity, however, are well documented: regardless of species, loss of genetic variety increases risk of genetic disease and loss of fitness. The following are known effects of low genetic diversity:
-
Increased rate of genetic disease
-
More sick days
-
Shorter lifespan
-
Increased susceptibility of autoimmune disease and infection
-
Smaller size
-
Smaller litter sizes
-
Decreased fertility of both bitch and dog
-
Increased puppy mortality
-
Decreased intelligence
It is worth noting, too, that designer dogs, mixed breed and random-bred dogs are not protected from the negative effects of low diversity by their lack of pedigree.

Interpreting offspring diversity in Litter Predict
The predicted litter average diversity is generated by comparing the specific diversity markers of the parents pair-by-pair to anticipate what the offspring are likely to inherit. It is not simply an averaging of the diversity percentages of the parents, which is why litters can have a higher degree of diversity compared to both their parents. For example, if a bitch and a dog are from very different, but low-diversity lines, when bred they may produce very diverse offspring. Each parent can only donate 50% of their total DNA to each of their offspring, so each puppy will have a different assortment of DNA from their parents due to genetic recombination. The actual diversity of each puppy will therefore vary depending on which particular marker combinations were inherited, reflecting what every breeder already knows: each dog in a litter is unique.
As good breeding is always to benefit the next generation, we list quick-to-read indications of the anticipated performance of the mating. Scientists agree that minimising loss of diversity is important, so breeders should aim to prioritise matings that at minimum produce puppies within breed average diversity, and preferably puppies that are more diverse than one or both of their parents, while also balancing other important considerations, such as disease carrier status and overall breeding merit.
How to avoid diversity loss
Certain breeding practices are at fault for the majority of lost genetic diversity in pedigreed breeds, but some historical events have also contributed. Read in more detail about leading causes of lost diversity to avoid some of the common pitfalls.
Frequently asked questions
What level of diversity loss is considered concerning?
There isn’t a specific line at which lost diversity starts to cause problems, as the risk is better described as an increasing level of likelihood with increasing levels of inbreeding. However, given most breeds experience some degree of inbreeding, some kennel clubs and geneticists have assigned guidelines or benchmarks for breeding recommendations. Some assign a COI value of 12.5% (equivalent to the breeding of two half-siblings from otherwise unrelated ancestors) or 6.25% (equivalent to the breeding of cousins) as the level to not exceed. The Swedish Kennel Club has additionally suggested that inbreeding (COI) should not exceed 2.5% over 5 generations, or 0.5% per generation maximum increase. However, we know from previous studies that the average COI of many breeds is around 25% or 33% heterozygosity in our testing, so additional planned inbreeding would far exceed the recommended COIs by most experts.
How is genetic diversity testing different from COI (Wright’s Coefficient of Inbreeding)?
Wright’s Coefficient of Inbreeding, abbreviated as COI, is an estimate of genetic diversity typically based on recent pedigrees and is associated with the probability of inheriting two copies of the same allele from the same ancestor. COI values are strongly influenced by the number of generations included in the calculation, and the completeness and accuracy of the pedigree. COI as a measure of inbreeding was invented in 1921 prior to our modern understanding of genetics and ability to efficiently test DNA.
Heterozygosity, or genetic diversity, is based on a comparison of inherited markers pair-by-pair from an individual’s sire and dam and the lineages they represent. Sometimes this is called “internal diversity” to distinguish from breed- or population-wide diversity. Heterozygosity testing is strongly influenced by which markers are chosen, their type, and to a lesser degree by the number of markers used. Genetic diversity testing for dog breeders was first made commercially available by Wisdom Panel™ in 2011.
Both methods of determining genetic diversity measure an association with inbreeding rather than directly measuring inbreeding itself.
How is your genetic diversity different from genetic COI (genetic coefficient of inbreeding)?
Coefficient of inbreeding (COI) by definition is based on pedigree, not genetic testing, but "genetic COI" is most often used to refer to heterozygosity adjusted for DNA segments known to lack variation within the breed due to recent or past inbreeding, called runs of homozygosity (ROH). This approach tends to give lower values for the amount of shared DNA between chromosome pairs due to this adjustment.
Our approach also measures genetic variation as heterozygosity and provides breed-based information about diversity norms, but these are based on tested breed averages, which also allows for accurate expectations of breed-specific diversity. However, because differing markers are used, and the two approaches use alternative methods for adjusting for breed-specific differences, the results are correlated but shouldn’t be compared.
Your percentage diversity is different than what I’ve received from other tests/companies. Why?
There is no standardised methodology for diversity reporting in dogs. Tests can differ in the following ways:
-
Calculation based on pedigree vs. genetic markers
-
Type of marker used (STR vs. SNP)
-
Use of breed-based adjustments (e.g. ROH)
-
Marker locations
-
Size and scope of breed database used for generating breed-based assumptions
-
Number of markers used
-
Method of reporting (Heterozygosity vs. Homozygosity)
All tests seek to approximate the level of actual inbreeding, but none are perfect. Studies have shown, however, that heterozygosity testing correlates well with coefficient of inbreeding and is not dependent on a complete pedigree.
I’ve tested full littermates and their reported diversity is different. Why?
Identical twins are extremely rare in dogs, so litters can be considered “fraternal." That means every puppy will be different, especially at the level of their DNA. Each parent can only donate half of their DNA to their offspring, and the particular segments of DNA passed down are subject to a process of genetic shuffling, called genetic recombination. This results in a spectrum of diversities within a litter, which will be even larger for dual-sire litters.
For example, Sibling A may have inherited a little more DNA from their dam’s sire (maternal grandfather), and Sibling B may have inherited more DNA from their dam's dam (maternal grandmother). If the grandsire and granddam on either the sire or dam’s side have very different lineages, you will see those differences appear as different heterozygosity percents, as heterozygosity is measuring the number of marker pairs that are different between the dam and sire. This is great news, as it allows breeders to maximize diversity in their matings to a degree not possible using pedigree-based COI alone, as COI calculations assume all offspring are identical.